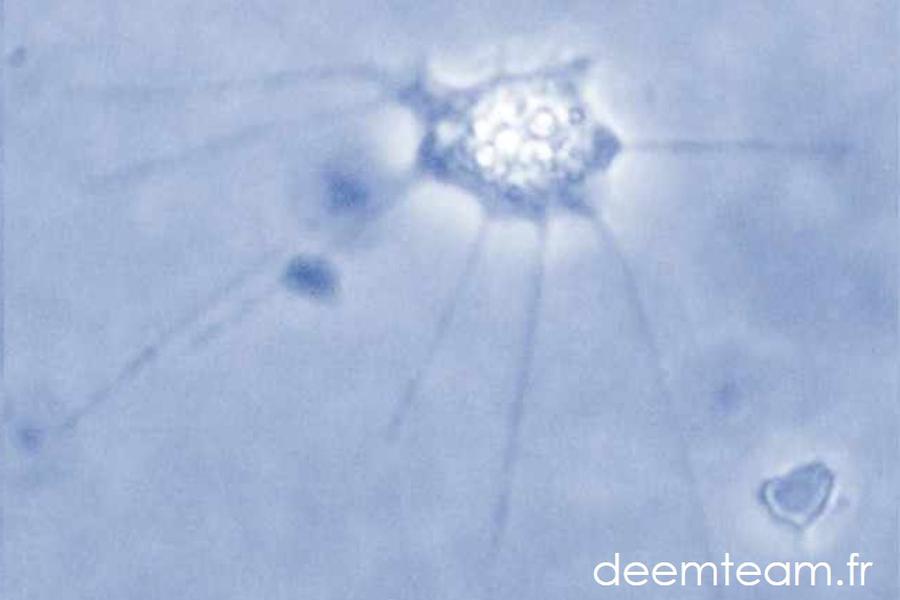
Microbial evolution cannot be understood out of its ecological context. We use molecular methods to explore the microbial diversity of prokaryotes (archaea and bacteria) and microbial eukaryotes in a variety of ecosystems. We are particularly interested in some extreme environments whose inhabitants are little known, and whose study could potentially lead to the discovery of novel or peculiar microbial lineages. We are currently working with samples from the deep ocean, multi-extreme environments, small freshwater systems, microbial mats and microbialites. Linking microbial diversity with mineral and physico-chemical data helps us identifying environmental determinants of community structure. We also study microevolutionary aspects related to microbial adaptation, specialization and biogeography. We also use environmental genomics (metagenomics) and single-cell genomics/transcriptomics to gain information about uncultured members of microbial communities from poorly studied ecosystems (e.g. deep-sea, microbialites, microbial mats).
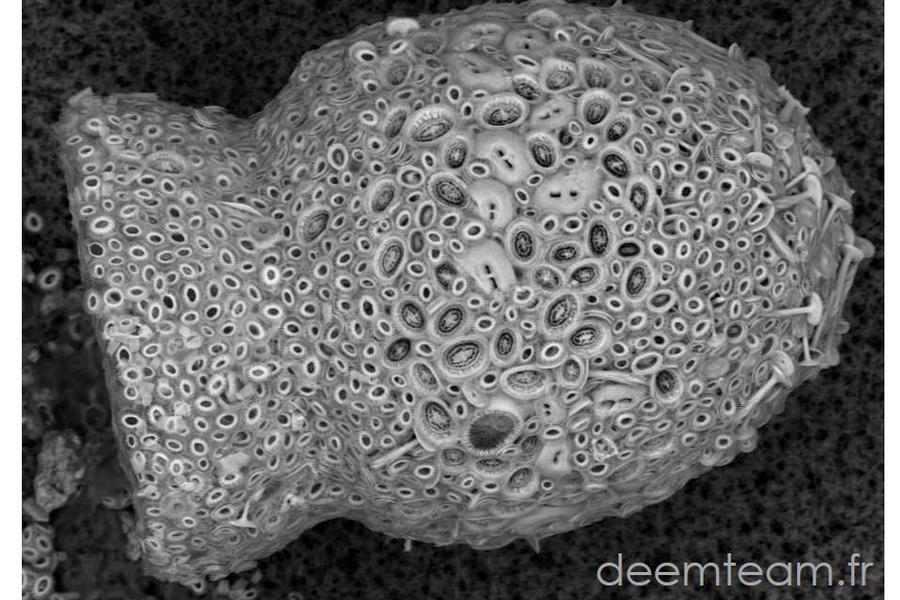

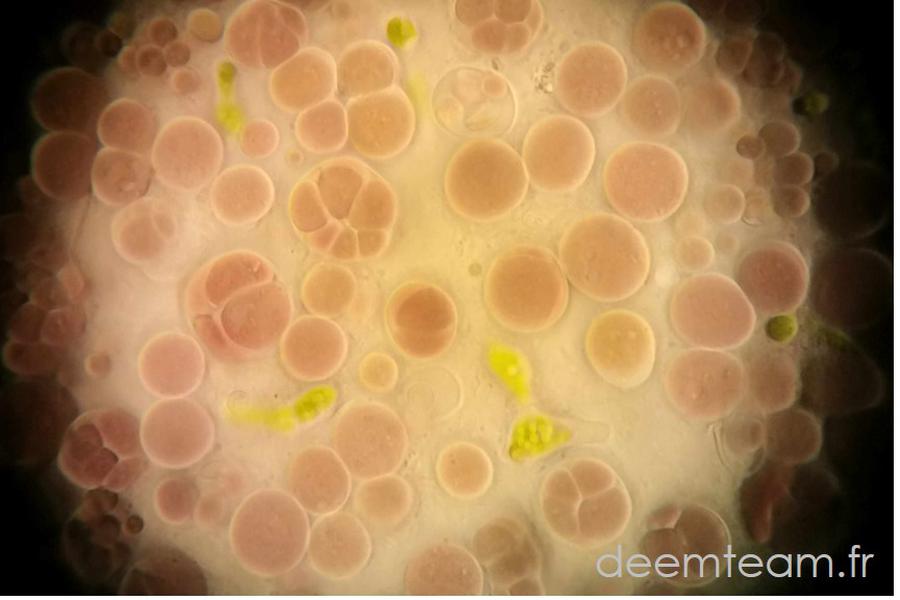
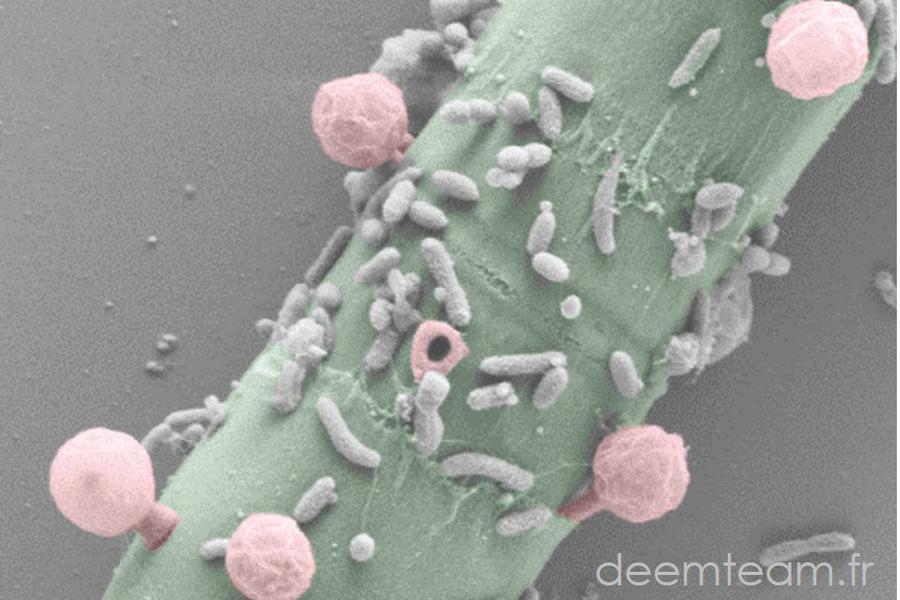
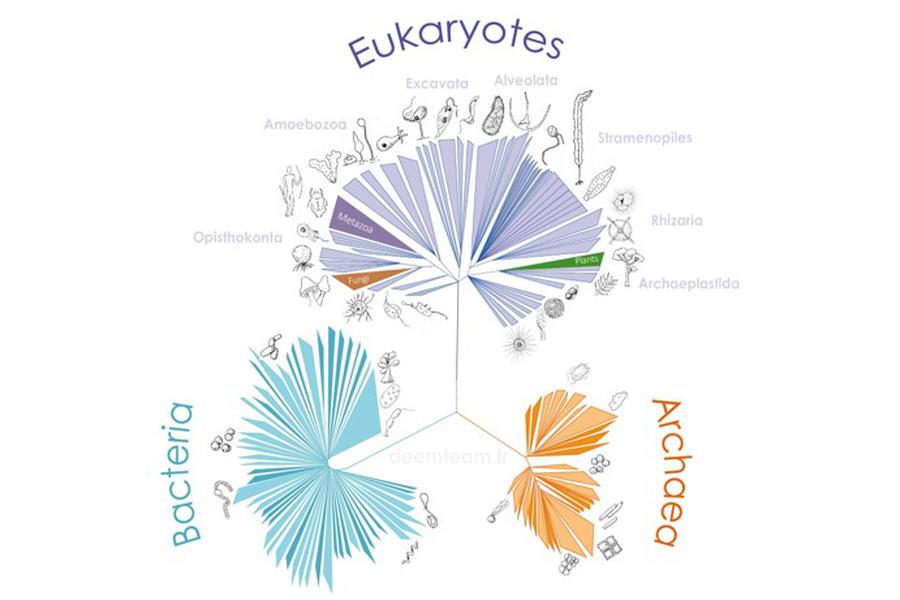
We carry out molecular phylogeny analyses using various approaches and molecular markers to try to solve problematic regions in the tree of life or to test evolutionary hypotheses. We also analyze environmental sequences that can significantly improve a taxonomic sampling that is often biased by parasite/pathogen sequences. Gene and genome information of uncultured lineages can be extremely useful for phylogenetic/phylogenomic analyses to address evolutionary questions such as deep phylogenetic relationships among lineages, key drivers of evolutionary diversification among major organismal lineages or the impact of horizontal gene transfer in evolution. The identification and use of environmental sequences with low evolutionary rates can limit the effects of long-branch attraction artefacts in phylogenetic reconstruction analyses.
We are interested in the function of lineages without cultured representatives, which can partly be inferred from metagenomic and single-cell ‘omic’ data. We are interested on how prokaryotic and eukaryotic communities assemble and vary in space and time as a function of biotic and abiotic parameters. DEEM is currently working on the ecology of microbial communities of some aquatic environments, in particular of small, shallow freshwater systems at the Paris region and the nearby Parc Naturel Régional de la Haute Vallée de Chevreuse. Due to their accessibility, it is possible to analyze temporal series and have a dynamic view of community structure through different temporal and spatial scales. Ludwig Jardillier’s group is involved in the study of carbon flux mediated by protists at the boundaries of the Southern Ocean. To this aim, he has participated in a research cruise at the Kerguelen islands and is funded by the ANR project Moby Dick.








Headed by Laura Eme and funded by the ERC Starting grant Macro-Epik [potential link with funded project page], this projects aims to understand how epigenetic mechanisms and LGT have shaped the macroevolution of eukaryotic genomes. Specific inter-connected objectives are